Tree-based Inference of Species Interaction Networks from Abundance Data
Abstract
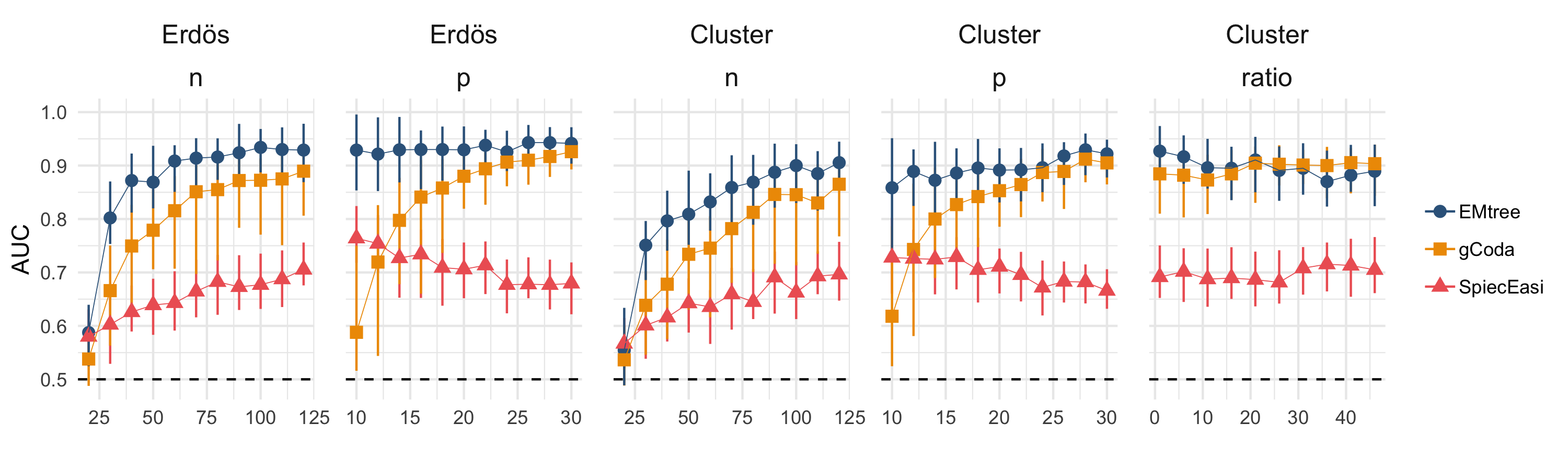
The behavior of ecological systems mainly relies on the interactions between the species it involves. In many situations, these interactions are not observed and have to be inferred from species abundance data. To be relevant, any reconstruction network methodology needs to handle count data and to account for possible environmental effects. It also needs to distinguish between direct and indirect interactions and graphical models provide a convenient framework for this purpose. We introduce a generic statistical model for network reconstruction based on abundance data. The model includes fixed effects to account for environmental covariates and sampling efforts, and correlated random effects to encode species interactions. The inferred network is obtained by averaging over all possible tree-shaped (and therefore sparse) networks, in a computationally efficient manner. An output of the procedure is the probability for each edge to be part of the underlying network. A simulation study shows that the proposed methodology compares well with state-of-the-art approaches, even when the underlying network strongly differs from a tree. The analysis of two data sets highlights the influence of covariates on the inferred network. Accounting for covariates is critical to avoid spurious edges. The proposed approach could be extended to perform network comparison or to look for missing species.