Resampling procedure for edges probability
ResampleEMtree( counts, covar_matrix = NULL, unlinked = NULL, O = NULL, user_covariance_estimation = NULL, v = 0.8, S = 100, maxIter = 30, cond.tol = 1e-10, eps = 0.001, cores = 3, init = FALSE )
Arguments
| counts | Data of observed counts with dimensions n x p, either a matrix, data.frame or tibble. |
|---|---|
| covar_matrix | matrix of covariates, should have the same number of rows as the count matrix. |
| unlinked | An optional vector of nodes which are not linked with each other |
| O | Matrix of offsets, with dimension n x p |
| user_covariance_estimation | A user-provided function for the estimation of a covariance |
| v | The proportion of observed data to be taken in each sub-sample. It is the ratio (sub-sample size)/n |
| S | Total number of wanted sub-samples. |
| maxIter | Maximum number of EMtree iterations at each sub-sampling. |
| cond.tol | Tolerance for the psi matrix. |
| eps | Precision parameter controlling the convergence of weights beta |
| cores | Number of cores, can be greater than 1 if data involves less than about 32 species. |
| init | boolean: should the resampling be carried out with different initial points (TRUE), or with different initial data (FALSE) |
Value
Returns a list which contains the Pmat data.frame, and vectors of EMtree maximum iterations and running times in each resampling.
Pmat: S x p(p-1)/2 matrix with edge probabilities for each resample
maxIter: EMtree maximum iterations in each resampling.
times: EMtree running times in each resampling.
Examples
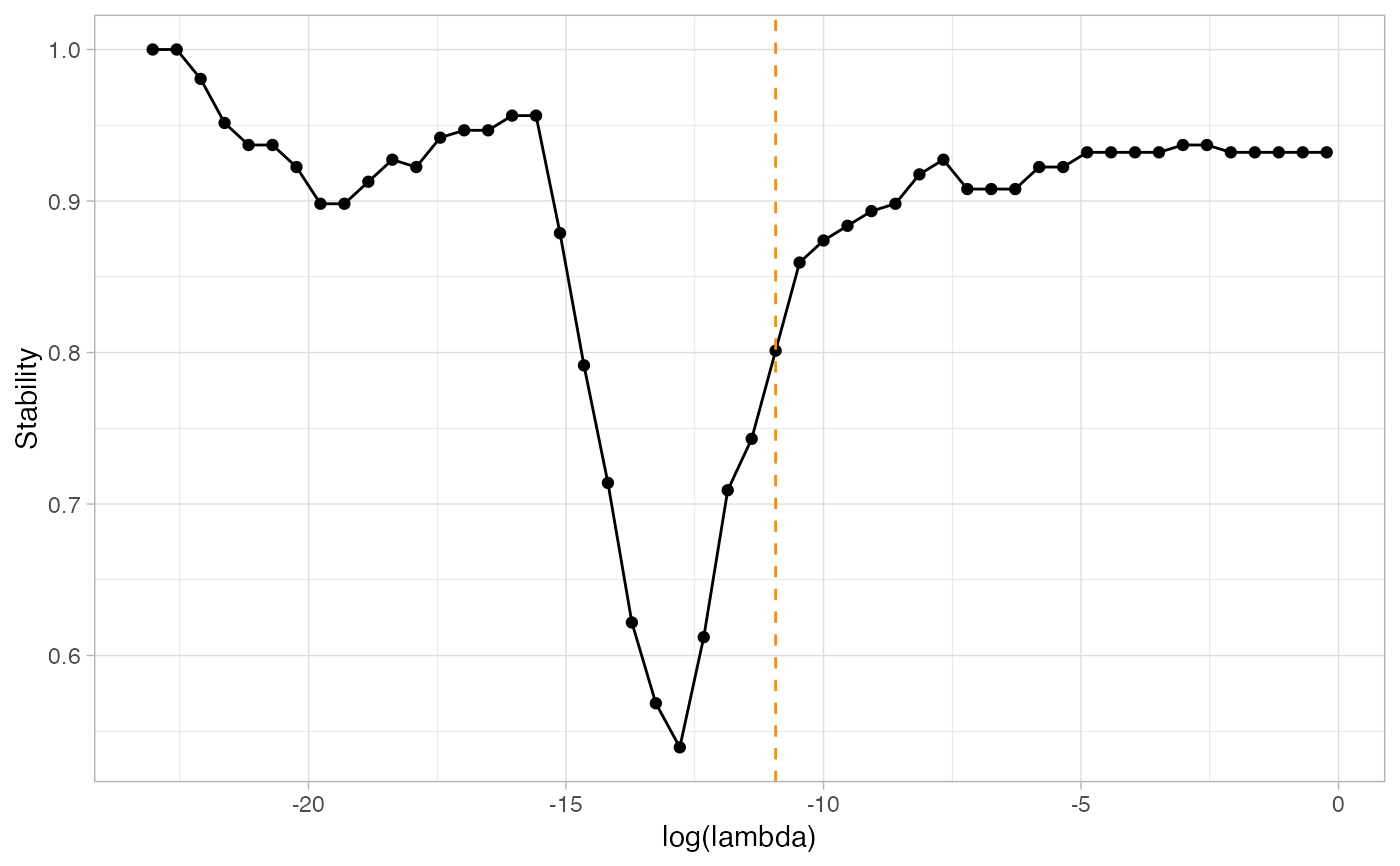
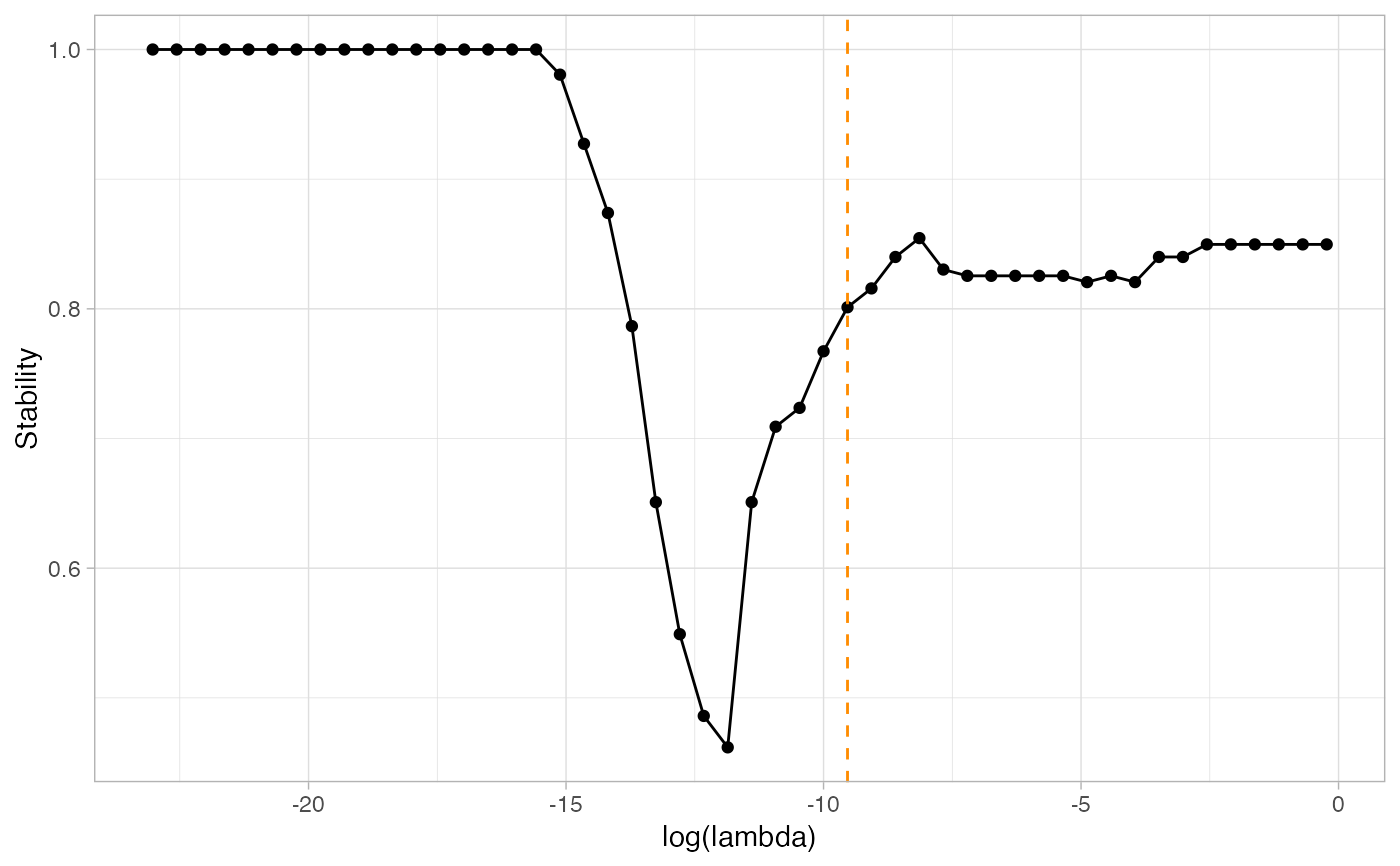
n=100 p=12 S=5 set.seed(2021) simu=data_from_scratch("erdos",p=p,n=n) G=1*(simu$omega!=0) ; diag(G) = 0 # With default evaluation, using the PLNmodel paradigm: default_resample=ResampleEMtree(simu$data, S=S,cores = 1)#> Computing 5 probability matrices with 1 core(s)... #> Convergence took 0.12 secs and 8 iterations. #> Convergence took 0.23 secs and 22 iterations. #> Convergence took 0.23 secs and 30 iterations. #> Convergence took 0.06 secs and 5 iterations. #> Convergence took 0.08 secs and 8 iterations.0.8 secs# With provided correlation estimation function: estimSigma<-function(counts, covar_matrix, sample){ Dum_Sigma = cov2cor(cov(counts[sample,])) } custom_resample=ResampleEMtree(simu$data,S=S,cores = 1,user_covariance_estimation=estimSigma)#> Computing 5 probability matrices with 1 core(s)... #> Convergence took 0.27 secs and 30 iterations. #> Convergence took 0.21 secs and 12 iterations. #> Convergence took 0.2 secs and 19 iterations. #> Convergence took 0.28 secs and 30 iterations. #> Convergence took 0.19 secs and 19 iterations.1.17 secs# We then run the stability selection to find the optimal selection frequencies, # for a stability of 85%: stab_default=StATS(default_resample$Pmat, nlambda=50, stab.thresh=0.8,plot=TRUE)#> truth #> pred 0 1 #> 0 50 1 #> 1 3 12#> truth #> pred 0 1 #> 0 49 5 #> 1 4 8